Example for Hyb-probe improvement using MeltCalc©
The ultimate thermodynamic modelling spreadsheet for Excel™
The following
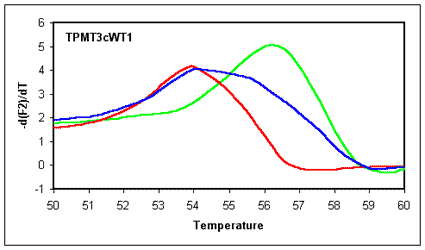
LightCycler™ melting curves have been achieved before and after thermodynamic
Hyb-Probe assay optimization using MeltCalc© Professional which runs
as Excel© "Add-In" in almost every Windows™ environment. Excel
7.0 or higher is required for execution. For this example
a 200 bp sequence surrounding the mutation of interest was given together
with the mutation
of interest (For details please read the
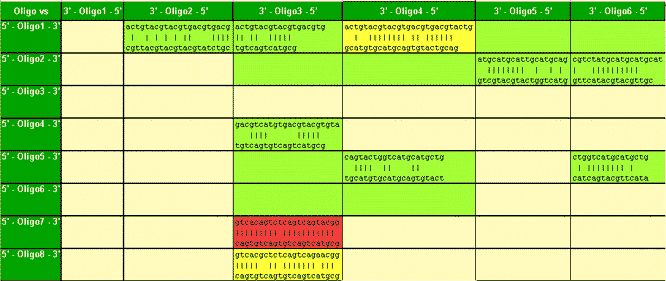
handbook). The program calculates 10 probe sets for each orientation
(sense and antisense) and each sequence (mutation or wild-type) and will
rank them according to the Tm-difference between mutation and wild-type.
The detection probe and a 3' and 5' adjacent anchor oligonucleotide are
calculated for each set. The program will by default ensure that the anchors
will have a >15% higher Tm than the matching probe. You can choose other
values on your own. Interested to get a fully featured license of MeltCalc©-professional?
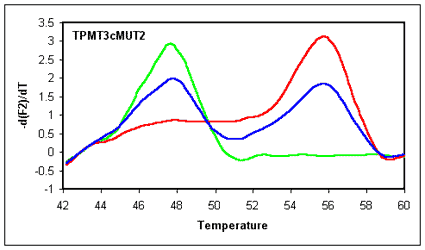
In the example shown both probes (wild-type and the mutation compatible)
hybridize with the antisense strand. The wild-type compatible probe (manual
selection) shows no sufficient Tm difference between perfect match and
mutation. However, the mutation compatible probe can reliably discriminate
the Wild-type and was automatically chosen by MeltCalc©.
The ultimate thermodynamic modelling spreadsheet for Excel™
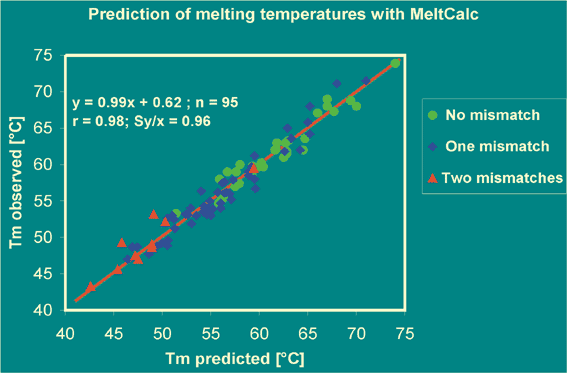
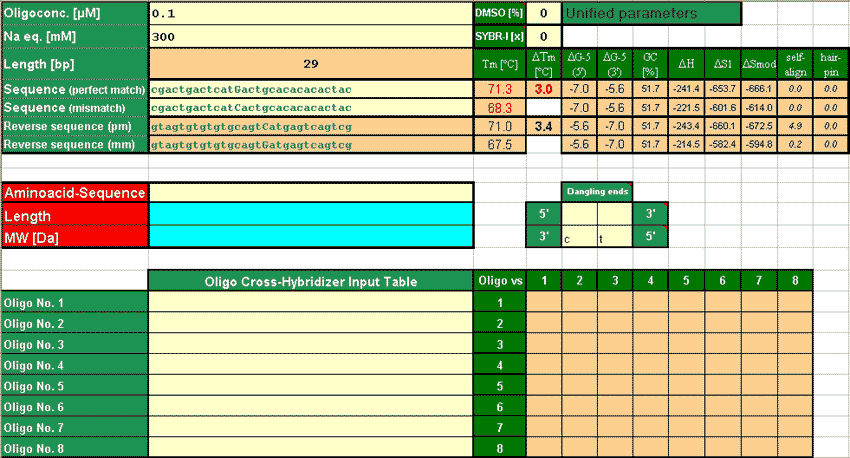
Calculation of Tm now including "dangling end"parameters (if applicable).
Maximum possible sequence length increased from 256bp to 1755bp for all features.
Maximum possible alignment for cross hybridization is shown graphically.
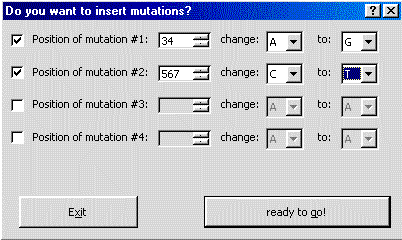
Mutations can be inserted by bp and base exchange.
A formula for calculation of Tm for long sequences is added .
Probe design:
Dangling end calculations are included into the algorithms.
(Highly important for optimization of hyb-probes; every probe has two dangling
ends, since the DNA template is usually longer than the probes!).
Space between anchor and probes for LightCycler™ assays can be chosen
(1 to 5).
Minor modifications:
Mismatches are highlighted automatically.
Calculation of sodium equivalents has been modified.
SYBR green-I™ influence on Tm is estimated, based on empirical data.
Cross-Hybridization Tms are highlighted using color coding in dependence of their hybridization temperature
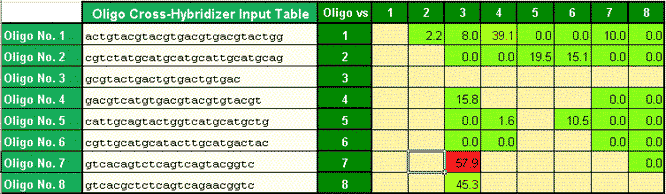
Cross-Hyb are shown graphically (partial display)